Target Prioritization: A case study on the BioBox Platform
How an early stage therapeutic company built a custom target prioritization system for renal cell carcinoma on the BioBox Platform.
Step 1: Defining Hypotheses and Organizing Data
Before utilizing the BioBox platform, the company addressed the following key questions to establish a clear direction:
What is our therapeutic approach?
What is our biological hypothesis?
What internal data will we work with?
What external data sources are relevant?
Answering these questions provided a foundation for the structure of their knowledge graph.
Step 2: Configuring a Custom Knowledge Graph
The core of the BioBox platform is a fully customizable knowledge graph. Think of the knowledge graph as a GPS for disease biology where the roads are relationships between destinations such as genes, pathways, variants etc. Explicit directions (data sources) tell you how you can get from point A to B.
Creating and maintaining a knowledge graph and the associated ontologies, is often a complex and time-consuming task. BioBox simplified this process, allowing therapeutic teams to focus on research rather than data wrangling.
Translational and computational teams mapped out the important biological relationships on the BioBox platform. The team started by loading preconfigured data packs from BioBox, which provided a robust initial framework. These data packs contained versioned ontologies and observations from consortiums such as OpenTargets, Alliance Genome, Clinical Trials and more.
They then uploaded their internal data, integrating all biological metadata with predefined concepts and relationships within the knowledge graph.
Wrangling ontologies, consolidating public knowledge bases and integrating internal data are processes that often take weeks to accomplish. Through the use of the intuitive UI and API we were able to accomplish this in a few hours.
- Director of computational biology
For more information on knowledge graphs and their role in drug discovery, refer to our detailed explanation here.
Step 3: Curating a Graph Model
A graph model maps important relationships between two concepts within the knowledge graph. For target prioritization, the concepts used were Genes and Diseases.
Each relationship, or "line of evidence," received a weighted score based on its importance. The team defined scientific criteria linking genes to diseases and categorized lines of evidence. Here are a few examples:
Gene Expression:
Upregulated genes from internal bulk RNAseq and scRNAseq data
Upregulated genes from TCGA bulk RNAseq data
Genomic Observations:
Variant frequencies in diseased populations (Internal data)
Variants that contribute to increase gene expression (OpenTargets)
Variants that are at risk/ protective for a disease (OpenTargets)
Epigenetic Information
Promoter regions that are hypomethylated in disease populations (methylation array (Illumina 850k)
Promoter regions there are acetylated in disease populations (H3K27ac ChIPseq)
Pathways and Biological Function:
Genes associated with specific pathways (Internal data + Reactome)
Genes that are markers for disease (Internal data + Alliance of Genome Resources)
Safety and Efficacy:
Clinical trial information, including trials with drug failures (ClinicalTrials)
Safety liabilities associated with genes (OpenTargets)
Drugs approved to target gene of interest (Chembl)
Negative scores were assigned to lines of evidence indicating poor target suitability, such as genes targeted by drugs that failed clinical trials. These scores were used to compute an overall weighted score for each gene, ranking them according to the team’s scientific criteria.
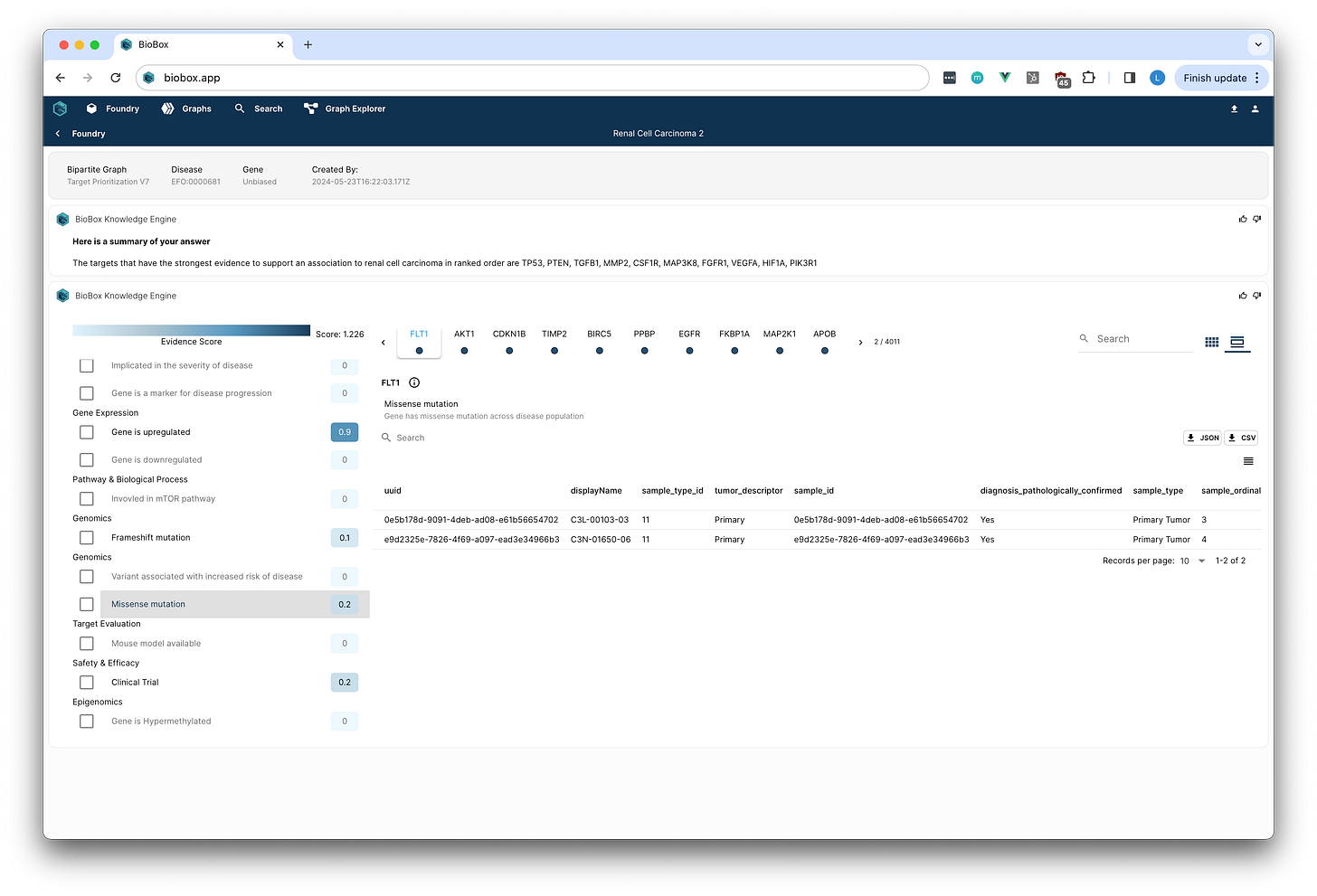
Step 4: Reporting & Ranking Targets
Once the graph model was curated, the team generated reports to evaluate targets. They selected "Renal Cell Carcinoma EFO:0000681" as the disease of interest and left Genes unbiased.
Within seconds, a report of genes ranked according to the scientific criteria that they curated was generated.
By selecting a target and a line of evidence scientists could see the supporting data identified within their knowledge graph.
The initial report highlighted well-characterized targets like BRIC5, AKT, and HIFa.
Using similarity and community detection algorithms, natively enabled in the graph model, the team was able to explore novel targets. Running a Jaccard similarity algorithm yielded targets that scored similar to well-characterized targets along the same lines of evidence. In other words, these targets might behave very closely to well-studied targets like AKT etc. This represented just one example of how the BioBox platform enabled principled and data-driven hypothesis generation through the exploitation of the graph data.
Based on the evaluation, the team adjusted weights and added additional lines of evidence to refine their prioritization system.
As new data was integrated into the knowledge graph, reports and target scores automatically updated, saving the computational team valuable time.
The impact of proprietary data on target prioritization was evident as the team compared reports from graph models with and without their internal data.
To further characterize a target the team explored all of its relationships within the graph explorer. A full break down on our graph explorer can be found here.
Conclusion
By leveraging the BioBox platform, the company efficiently built a custom target prioritization system for renal cell carcinoma. The comprehensive integration of internal and external data sources, coupled with BioBox's customizable knowledge graph and robust analytical tools, provided several key benefits:
Time Savings: The streamlined ontology management and automated data integration allowed the team to quickly get started and maintain momentum, reducing the time typically spent on data wrangling.
Cost Efficiency: By efficiently organizing and analyzing data, the company was able to focus resources on the most promising targets, reducing unnecessary expenditures on less viable candidates.
Enhanced Confidence: The weighted scoring system and robust evidence framework gave the team greater confidence in their target selection, supported by clear, quantifiable data.
Comprehensive Evidence: The inclusion of diverse lines of evidence, such as gene expression, genomic observations, and clinical trial data, provided a thorough understanding of each target's potential, ensuring well-informed decision-making.
Cross-Functional Collaboration: The platform’s collaborative features enabled seamless communication and data sharing among team members, fostering a cohesive and efficient working environment.
Note: Proprietary data has been redacted for privacy. Sequencing-based observations in images above have been derived from TCGA data.
About BioBox
BioBox is a data intelligence platform for drug discovery. Rapidly build a custom data graph and deploy graph ML models for target prioritization, indication selection, and MOA analysis. Teams using BioBox make faster and better data driven decisions to de-risk drug programs and get assets into clinical studies quicker.
To learn more about how BioBox can supercharge your discovery pipeline, book a demo with one of our team members!