Beyond Data: The Rise of Knowledge Graphs in Accelerating Drug Discovery
We are now living in the era of biological big data. The cost of sequencing is rapidly decreasing. Simultaneously, we are witnessing the rise of bio-computing tools and platforms that enable scientists to process sequencing data at remarkable speed and scale. This abundance of data, when harnessed effectively, holds the key to making informed decisions around target identification and validation stages in drug discovery. Selecting a good target to drug is one of the earliest and most consequential decisions made when developing new therapeutics.
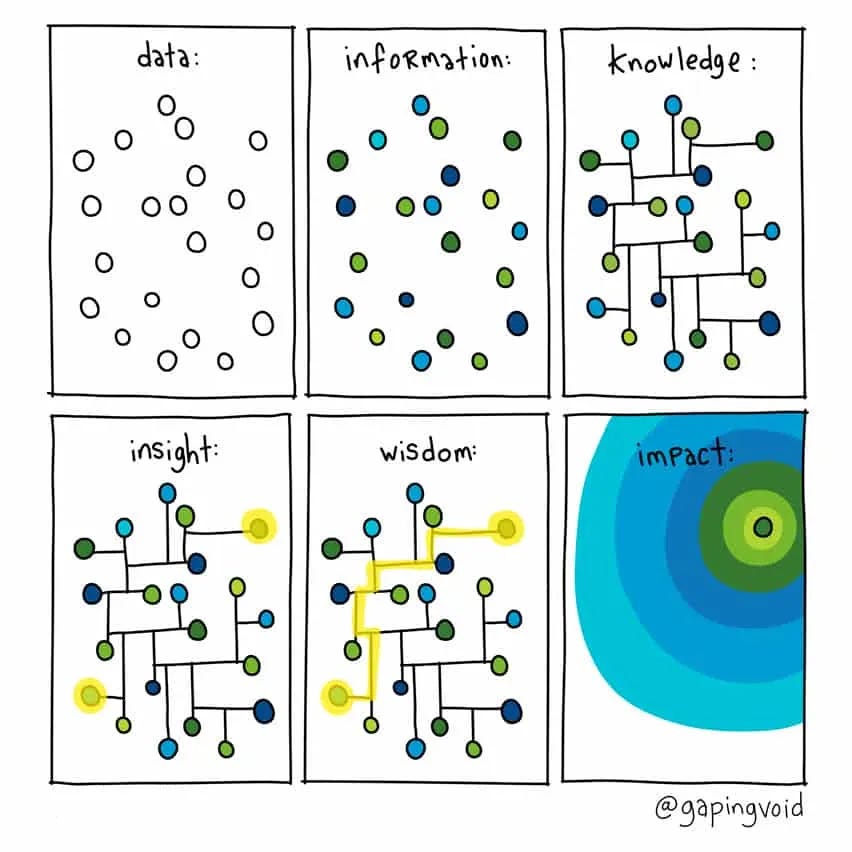
Today, the challenge often lies not in the acquisition of data, but in its interpretation and utilization. Simply having data is no longer enough. To fully exploit the volume and variety of data available, we need systems to capture the relationships and patterns from observations to form a more holistic view. This is where Knowledge Graphs (KG) truly shine, and when properly used, will completely transform that way data is used in drug discovery. In this article, we describe at a high level, what a Knowledge Graph is and how to get started crafting one.
What is a Knowledge Graph?
At its core, a Knowledge Graph (KG) is a formal structure designed to represent information as a set of entities and the intricate relationships between them. In simple terms, we can think of individuals like BRCA1 or Breast Cancer as nodes, and relationships like mutated_in as descriptive edges between nodes.
When developed properly, it can enable automated reasoning and inference to unearth hidden implicit connections that often belie breakthroughs and discoveries. Many leading biotech and pharmaceutical companies have been ingesting data and constructing internal knowledge graphs to use in a variety of applications from drug repurposing to target identification.
DeepLink, Janssen Pharmaceuticals: Using DeepLink, an internal knowledge graph platform, researchers successfully identified two hallmark targets (one of them is now in their portfolio) for pulmonary hypertension.
ARCH, AbbVie: ARCH is the name of AbbVie’s internal knowledge graph. In a case study, AbbVie scientists used the embedded logic to discover a putative therapeutic in their portfolio that could be used to treat Carney Complex, a rare and deadly disease with no approved treatments.
The essential and most important component of a KG is the underlying ontology. Ontologies are semantic data models which determine the types of concepts that exist in the KG. Importantly, they are distinct from individual (data points) in the KG because they represent entire categories of concepts, not a specific named concept. For example, instead of describing the gene SOX2 and specific properties about SOX2, the ontology focuses on defining the concept of Gene and capturing the characteristics that a Gene should have. Some of these characteristics can be relationships to other concepts in the ontology. For example, we can describe the relationship between a Gene and Pathway with participates_in.
Using this ontology, data can be structured in an interpretable way, and forms the KG.
Tangible Benefits: Drug Repurposing
For biotechnology or pharmaceutical companies that have existing assets in their portfolio with a defined target and mechanism, we can expand the market value of the asset if we can find alternate use-cases for it.
Suppose your asset BX123 selectively inhibits the G1 gene and is an approved treatment for Disease Y by rescuing an overactive pathway P1. When enough data is modeled into a knowledge graph, we can start to make some inferences and uncover new insights. For example, it is also known that P1 activates transcription of G2, which in turn, activates P2 that is implicated in the etiology of Disease X. Through walking the KG, it can be inferred that the asset BX123 could be a viable treatment for Disease X, expanding the use-cases for the asset and ultimately the market value.
Tangible Benefits: Target Identification
KGs can also help researchers make informed decisions when selecting a target to drug. Using sequencing datasets, we can load in observations such as gene expression, differential expression, mutations etc. to enrich the knowledge graph. For example suppose we have normal tissue expression data and disease-specific patient tumor sequencing data.
At first glance, G1 and G2 appear to be viable targets as they both participate in an overactive disease-causing pathway, but after considering information loaded from sequencing data, G2 seems to be most viable. Here’s the reasoning:
G2 is selectively upregulated in disease
G2 is poorly expressed in normal tissue
G2 is significantly upregulated in Disease X compared to healthy normal controls
G1 is not contributing to overactive pathway
G1 is abundantly expressed in normal tissues and not found to be differentially expressed in Disease X. It stands to reason that if G1 is implicated in Disease X, all brains would have Disease X.
In this hypothetical scenario, it illustrates how the incorporation of observations from sequencing datasets into the knowledge graph enables researchers to conduct evidence-based reasoning quickly and effectively.
Challenges and Considerations
Despite the tangible benefits that KGs can bring into a biotech research team to accelerate drug discovery, there are significant challenges to making the KG effective. Here are some considerations to make if you are thinking of starting a KG initiative.
Build a shared dictionary
Don’t be an ontology goblin.
The goal is to have shared knowledge. The ontology should be built with all stakeholders, from computational biologists to business teams, in mind. Successful companies implementing KGs have a common understanding of how things are defined using a shared vocabulary in their organization. This massively reduces communication errors and makes data ingestion, harmonization, and reporting a breeze. It matters not where this is done, but that it is visible and available to everyone.
Connect data streams
Before committing to building a KG, be sure to plan out where the data comes from and if it can be reliably and efficiently connected to the KG. Building the perfect ontology and infrastructure is meaningless if you cannot consistently add data to the graph. While they can vary by use-case, the most common ones would be LIMS metadata, bioinformatic workflows and outputs, ELNs etc.
Bottom Line
Knowledge Graphs are tools that can drive massive acceleration in the drug discovery process. There is growing adoption and use of knowledge graphs in big pharma and biotech. When harnessed effectively, it enables research teams to reason over the data at scale and unearth hidden patterns.
However, effective KGs require significant time and capital resources, which may limit adoption in early-stage startups and biotech companies, despite the immense value KGs hold. This is where purpose-built platforms like BioBox can help. We empower smaller research teams with dedicated tools to:
collaboratively build ontologies, with access controls and versioning
integrate with data streams to power your knowledge graphs
explore and mine knowledge graphs for insights
Ready to get started? Book a Demo